Dr. Giedrė Tamulaitienė – about Protein Structures, Their Beauty and Why It Is Beneficial for Researchers to Collaborate with Artists
The structure of a protein is like a photo of it, allowing you to take a closer look at the world of molecules. And researchers have to work hard to see that. Structural information about the proteins is also very important in the field of bacterial antiviral defense systems, the study and application of which have already been awarded the Nobel Prize twice – for gene and genome editing tools. The latter is already used in Europe for the treatment of sickle cell anemia. We currently know more than 100 bacterial antiviral defense systems, which were studied and discovered by the aforementioned tools. “Only a few of them have been characterized. Therefore, we do not yet know what we will find and how researchers will come up with ways to apply them”, says structural biology researcher Dr. Giedrė Tamulaitienė of Vilnius University (VU) Life Sciences Center (LSC).

Dr. Giedrė Tamulaitienė
Dr. G. Tamulaitienė has been working in the field of structural biology for more than 20 years. Her research has recently been published in the prestigious journals Science and Nature. The researcher not only determines the structures of proteins but also creates pictures inspired by their beauty: one of them could be seen at the MO Museum exhibition in 2019.
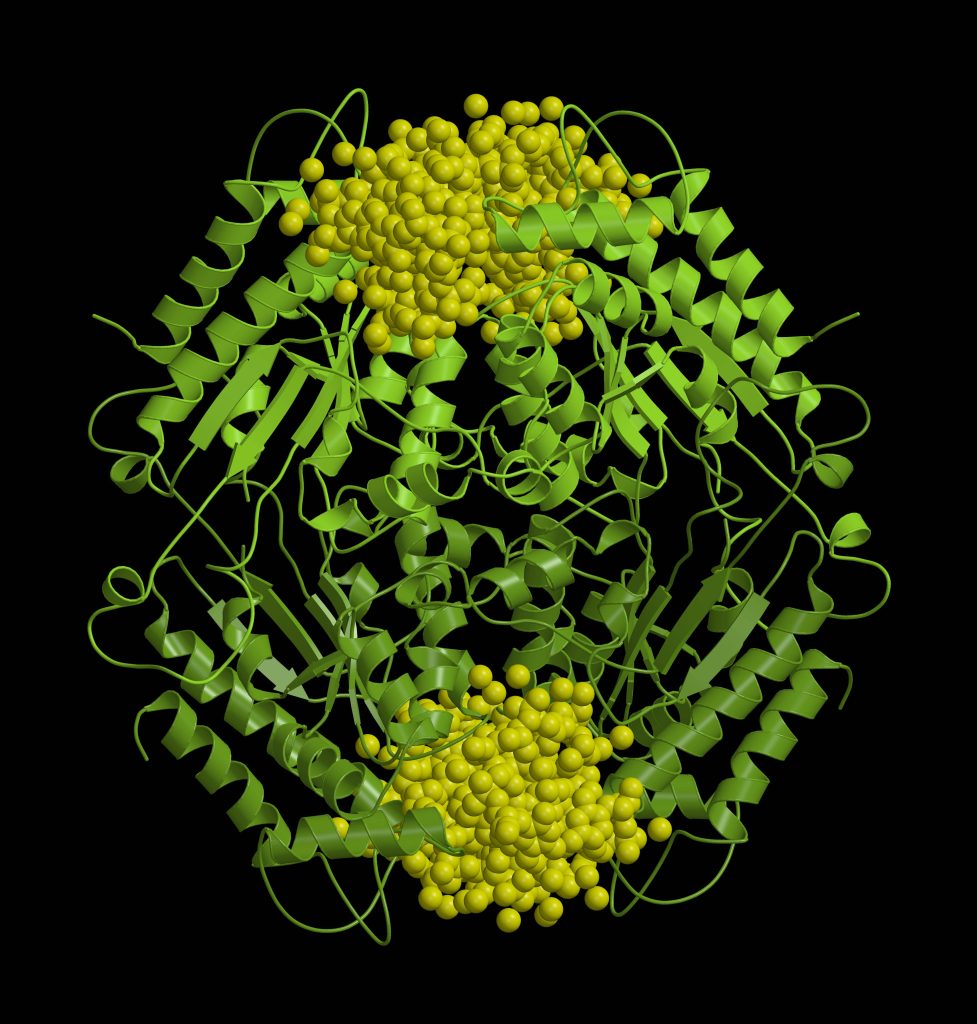
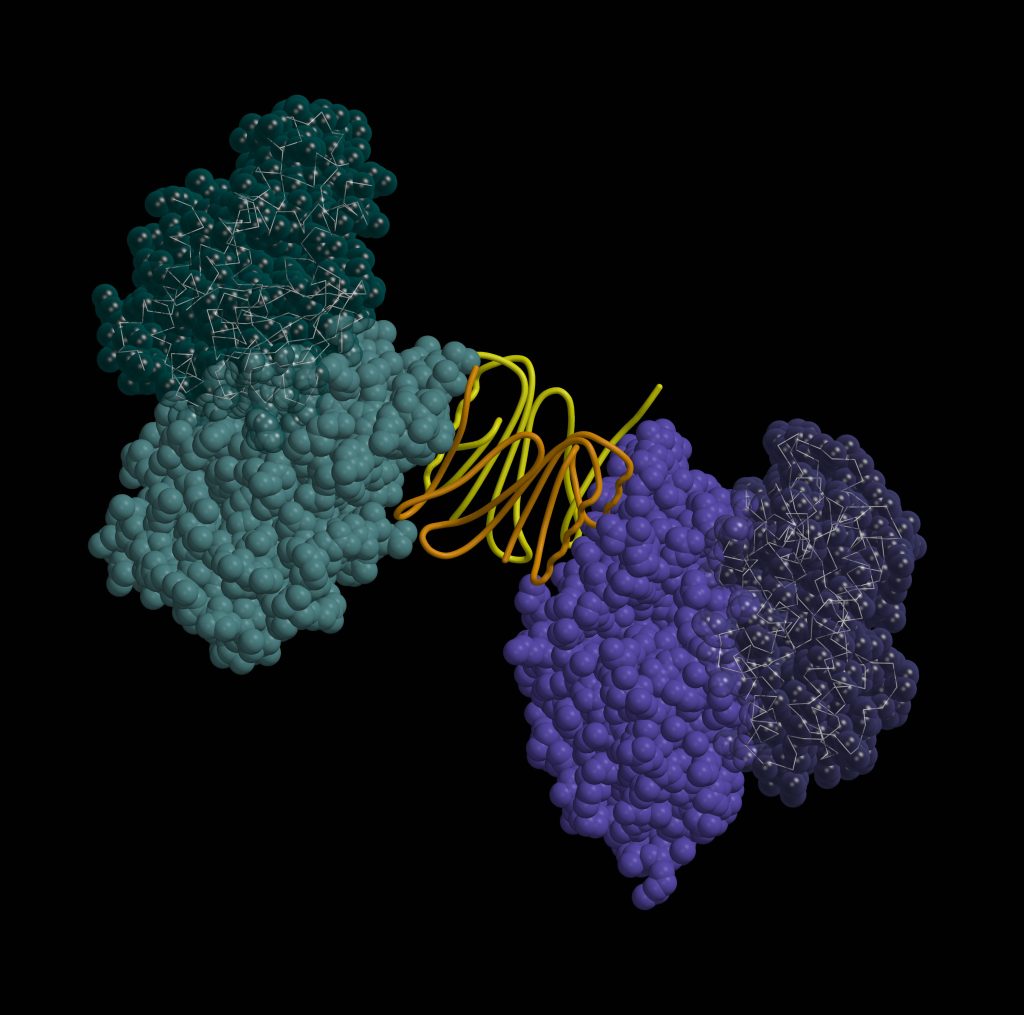
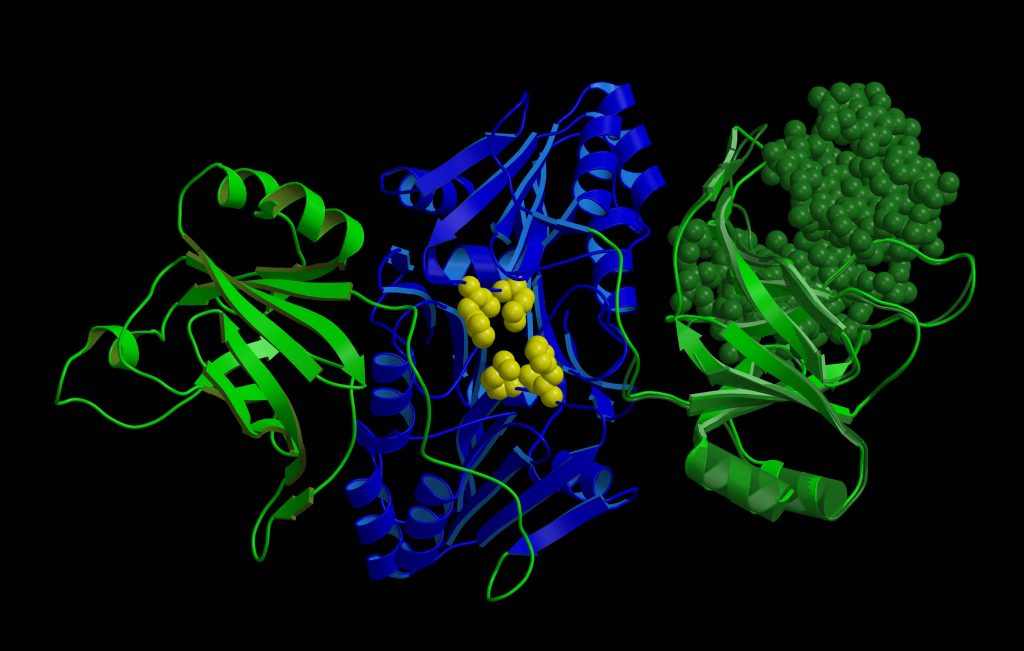
“We have a saying that the goal of structural biologists is beautiful pictures (laughs). Indeed, one of the most enjoyable moments in the work is the birth of the protein structure. I start with the experimental data, such as three-dimensional maps electron density clouds, and I begin to model the structure of the protein into them. I rotate the amino acids on the computer screen so that they fit in the density map as accurately as possible. I understand how they should be placed, and that’s how the structure that we then see in the pictures is born little by little. It is an extraordinary feeling to discover a new, previously unknown small molecule, unexpected protein interactions or complex protein helical structures in that new structure,” Dr. Giedrė Tamulaitienė starts the conversation.
How long does it take to build up an image of the protein structure?
Several methods are used to obtain these images. We use X-ray structural analysis and cryogenic electron microscopy.
For example, in the case of X-ray structural analysis, the gene of the protein of interest first has to be cloned into the selected expression system so that the bacteria produce a lot of the protein, then the protein is purified and concentrated. And then the process of structural biology begins: first of all, you have to get protein crystals. This can take a very long time because you have to try endless different conditions. We use crystallization robots to facilitate protein crystallization.
When we get a crystal, which is usually less than a millimeter long, we expose it to X-rays and obtain a diffraction image. We then use synchrotron radiation to collect a diffraction dataset. According to the positions of the spots and their intensity, we obtain a map of the electron density with the help of a number of computer programs. Then we use the obtained map to model the structure of the protein. This process really usually takes quite a long time, from several months to even several years.
From left to right: Mimosa, Unfulfilled Dream, Choice
You get the structure of a protein, what’s next?
You could say that the structure of a protein is like its picture. Only it is not two-dimensional but three-dimensional. We can zoom in, and look at it from all sides, compare it with the structures of other known proteins. We have a suspended moment in the “life” of that protein. We can judge whether a protein is mobile or flexible. We also can predict which parts of the protein are important for its specific function.
How do we know that a part of a protein is important for its function? The easiest way is to simply damage that part of the protein. We introduce the so-called mutations. If we manage to “damage” the protein as expected, then we have guessed the function of that part.
What are these proteins?
I mostly study proteins that belong to the antiviral defense systems of bacteria. These proteins help bacteria to defend themselves against viruses (called bacteriophages) that attack them.
And why are you studying these particular proteins? As far as I know, it is research like this that led to the discovery of genome editing tools like CRISPR-Cas9.
Yes, exactly. You could say that I came to this field by accident. When I was looking for a place to do my internship while studying at Vilnius University, I came across the laboratory of the Institute of Biotechnology, where they were looking for new enzymes of the antiviral systems of bacteria, restriction endonucleases belonging to the restriction-modification systems. At the time, it was one of a few well-known and studied bacterial antiviral systems. Research on restriction endonucleases has been of interest mainly because of their great applicability as gene-cutting tools.
Somewhat later, other systems were discovered, the most famous of which is the genome editing tool you mentioned, CRISPR-Cas9. Two Nobel Prizes have been awarded for the discovery and application of gene and genome editing systems.
What applications do you think new knowledge about bacterial antiviral defense systems could lead to? I understand that your research is fundamental, so predictions like this are difficult, but in terms of CRISPR-Cas9, just recently the treatment of sickle cell disease using this tool has been approved in the UK and the European Union.
For science, the discovery of CRISPR-Cas9 was very important for genome editing in living cells and more complex organisms (plants, animals) as well as for the creation of transgenic organisms. Transgenic animals are created for scientific purposes to obtain information about gene function and regulation, including human diseases, and to obtain therapeutic products, recombinant pharmaceutical proteins, for use in the treatment of people. CRISPR-Cas technology has made these methods much faster and cheaper. Also, during the COVID-19 pandemic, new diagnostic tools have been developed based on various CRISPR-Cas systems.
Currently, more than 100 bacterial defense systems have been discovered, and only a few of them have been characterized. Therefore, we do not yet know what interesting things we will find and how researchers will come up with the application of these systems.
Theoris Spiral
What work are you most proud of?
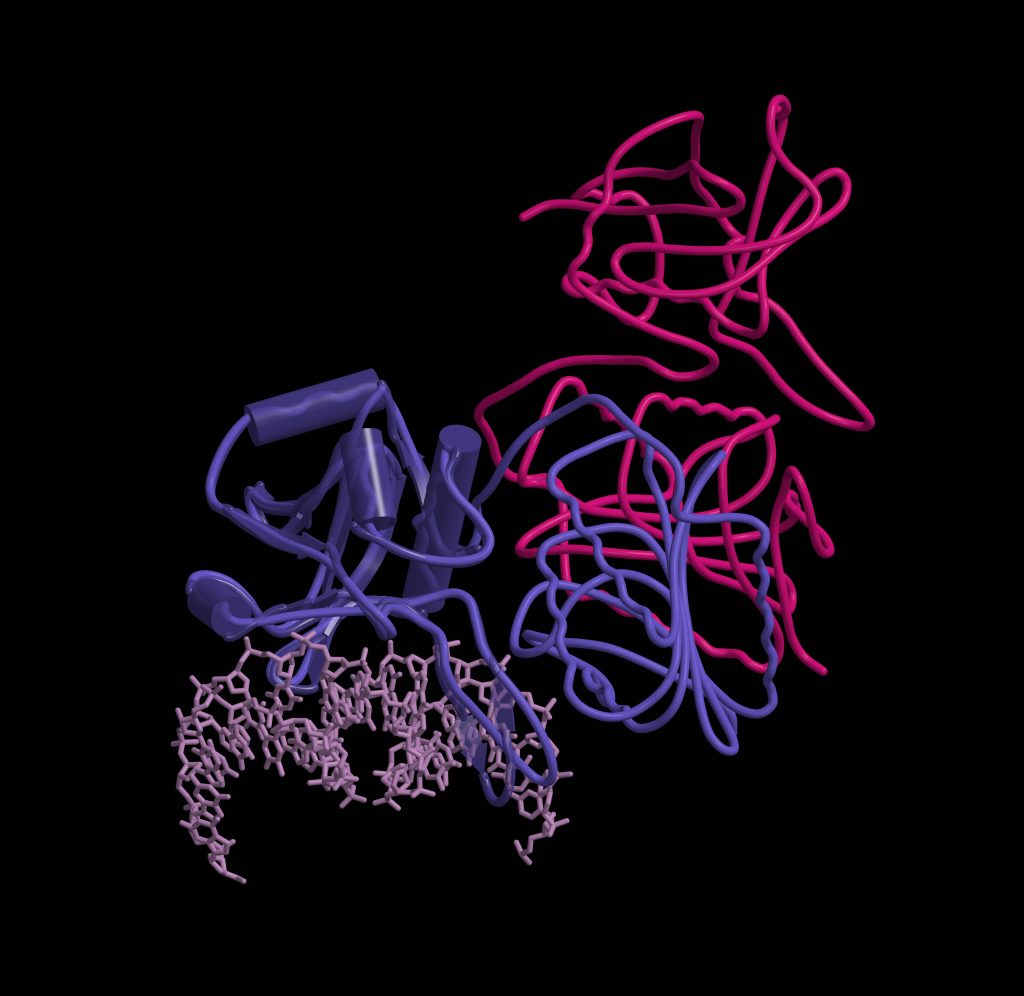
I am most pleased with my research in a project funded by the Research Council of Lithuania (RCL), which is dedicated to the research of the bacterial antiviral defense system Thoeris. In the project, we analyzed the bacterial defense system Thoeris, named after the Ancient Egyptian goddess of childbirth and fertility. This and 8 other systems were discovered by Israel researchers in 2018, naming each of them after their guardian deities. Interestingly, one defense system received Gabija’s name, marking the contribution of Lithuanian researchers in this field. When studying the Thoeris system together with my team, we found that when a virus attacks a bacterium and the Theoris sensor protein produces a signaling molecule, another protein of this system binds this molecule and is “turned on”: it forms a long helix-shaped thread, a filament, which destroys one of the essential molecules for the cell. In this way, the bacterium “kills itself” and prevents the spread of the virus. A study describing this helix was published in the prestigious journal Nature earlier this year.
You are one of the few scientists who can boast of an exhibition. In 2019, you organized the exhibition Structural Stories, you gave a public lecture at the MO Museum about the beauty of protein structures and how they inspire artists.
Well, proteins are just very beautiful to me… I really like making both scientific illustrations and artistic pictures of structures. In the beginning, it was just a hobby. When we collected a lot of images of structures in our department, the head of the department, Prof. Virginijus Šišknys, suggested using them to make a calendar. I agreed to do it and I really enjoyed it. It was in 2009. That’s a long time ago (laughs).
After that, we also thought about hanging pictures of structures on the walls in our department. Agnė Grinevičiūtė, a communication specialist at LSC, saw our pictures and suggested making an exhibition of them. This is how the exhibition in the LSC was gradually born. At the time, the Center was visited by Prof. Viktoras Butkus, one of the founders of the MO Museum, who found one of my pictures similar to a new painting in the collection, whose abstract forms resembled protein structures. Thus, one of my works Choice was exhibited in a large format in the MO Museum.
From left to right: Spring, Thoughts
In the lecture, you look at the creators of visual arts who are inspired by protein structures. Can you name a few of them?
There are not many such works in Lithuania. Usually, such protein sculptures decorate large research centers and hospitals. Artists try to reveal not only the beauty of proteins but also their deeper meaning: for example, German sculptor and physicist Julian Voss-Andreae, who lives in the USA, “unwraps” the protein collagen structure and the sculpture becomes a metaphor for aging. Julian Voss-Andreae’s sculpture of hemoglobin, the oxygen-carrying protein in the blood, resembles a steel heart. Canadian painter Jacques Deshaies has said that proteins are as beautiful to him as flowers.
Do you think researchers and artists can help each other?
Definitely, illustrations of scientific results created by artists can help other researchers and the public to understand the discoveries, what the researchers wanted to say, what they discovered, and present it in a very artistic way.
Art can help bring science closer to society. People sometimes think that science is so complicated that it’s not even worth trying to understand. Observing the work created by the artist, everyone has the opportunity to find their own relationship with science and feel the beauty of nature.
I think art inspired by science can be used very well as a teaching tool. For example, pupils can create joint projects together with biology and art teachers. This would enable children to “touch” science. Protein Data Bank has prepared a project for teachers with detailed methodologies on how to help children start liking science through protein structures. Perhaps after such a project, children will someday want to determine the structures of proteins themselves.
Thank you for the conversation.
Thank you too!
Interviewed by Goda Raibytė-Aleksa